Creating the Trends in Genetics October 2012 Cover
contents
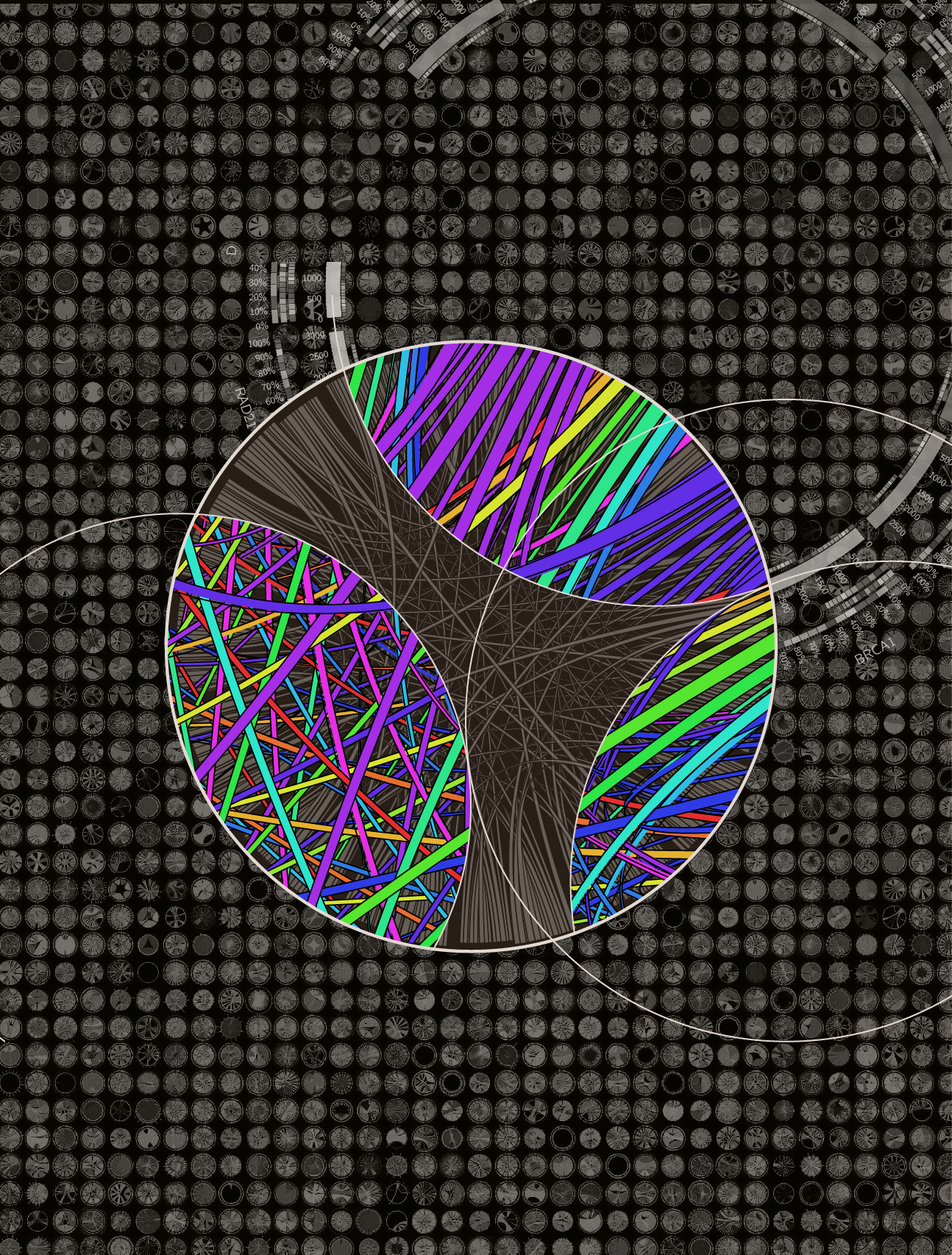
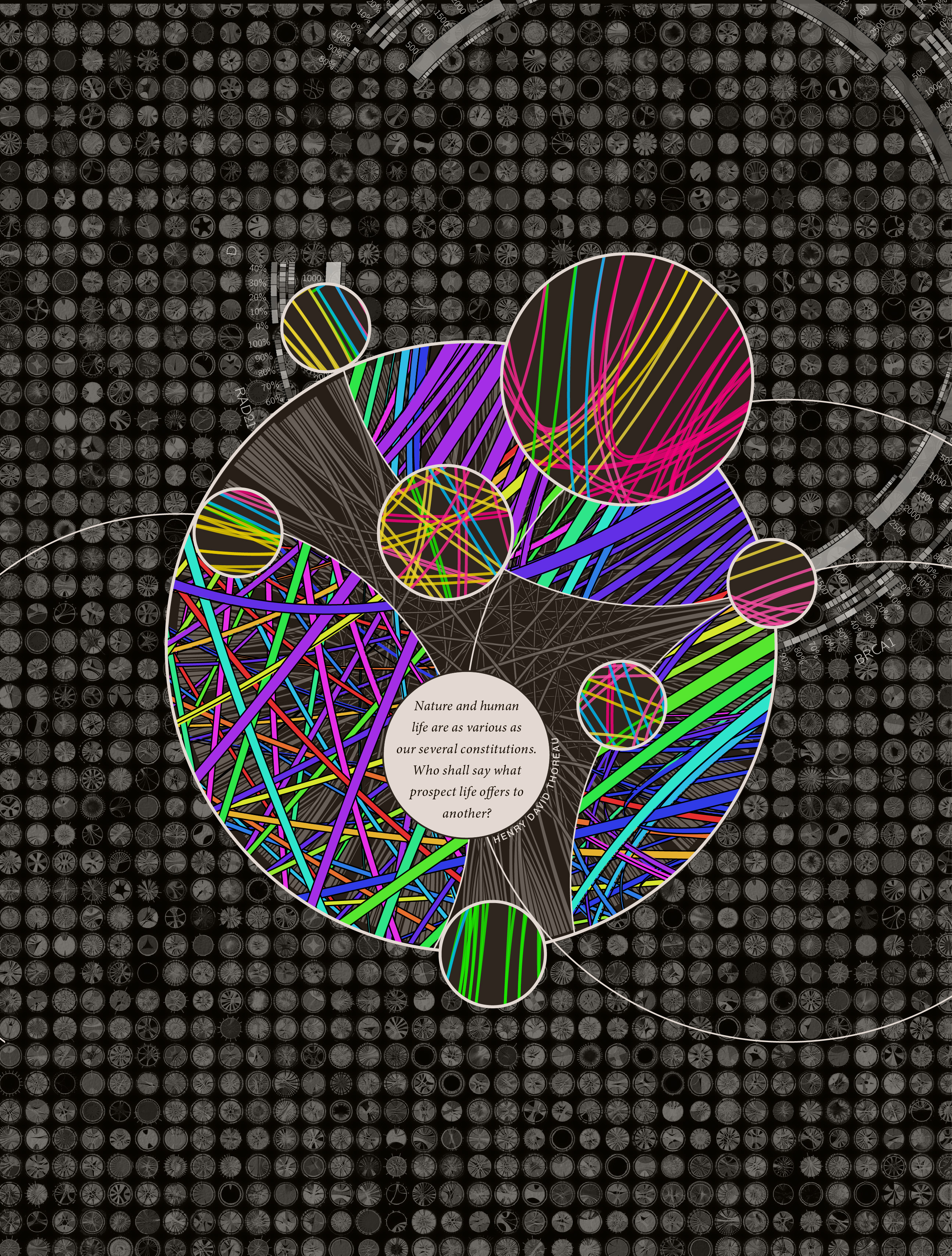
Lately, I've been making a lot of square things round. So when Rhiannon Macrae, the Editor of Trends in Genetics, requested a Circos-like cover image for the human genetics special edition of the journal, I started drawing circles.
The image was published on the cover of Trends in Genetics human genetics special issue (Trends in Genetics October 2012, 28 (10)).

Browse my gallery of cover designs.


I have a collection of unpublished Circos posters and thought these might be a good starting point. Rhiannon and I narrowed the choice down to the black-and-white design that showed sequenced organisms. We also liked the complex style of a panel of hundreds of Circos images generated with the tableviewer.
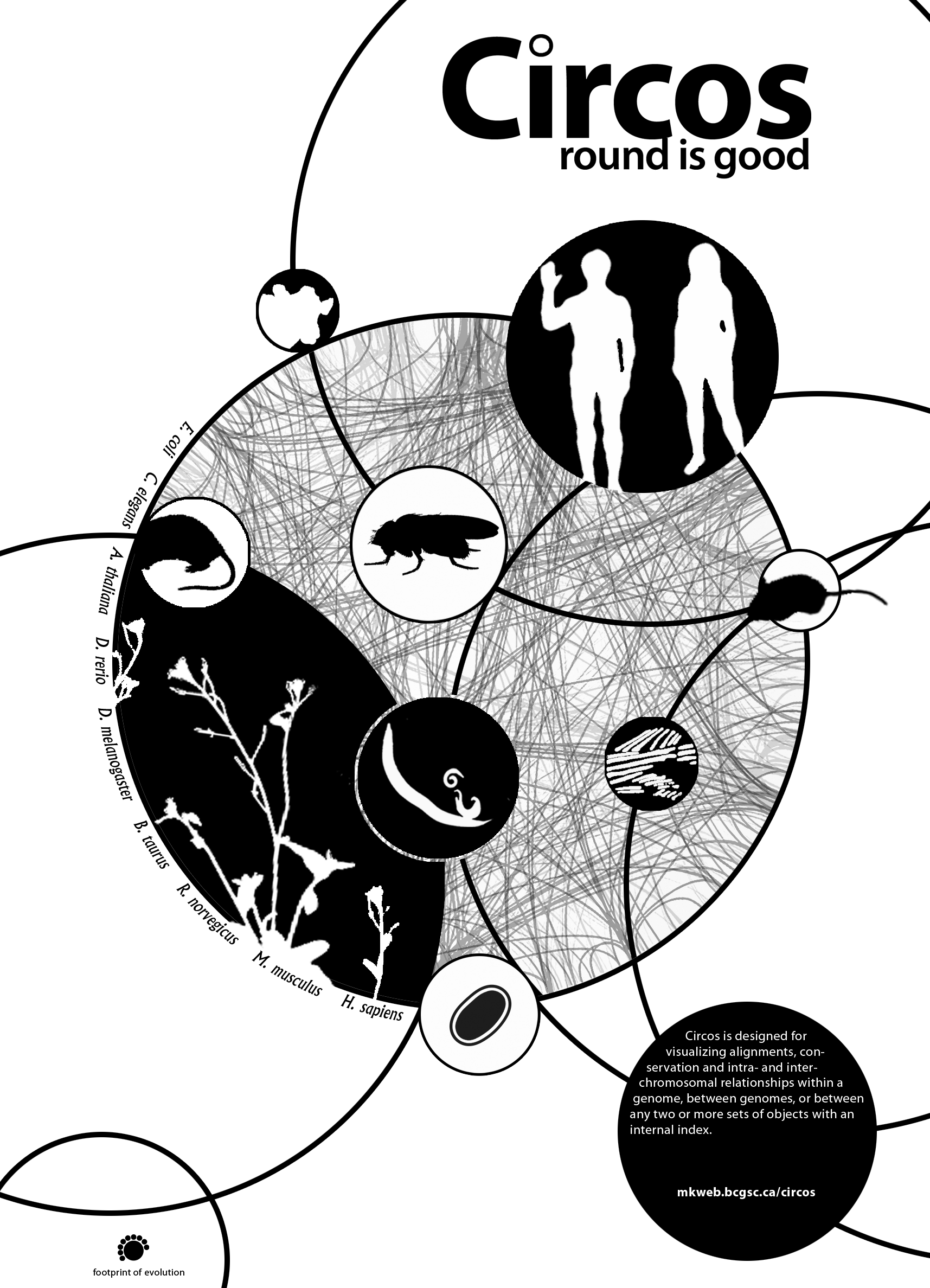

The idea would be that the foreground would be more artistic and stylized, while the background was more technical and complex. I have thousands of images available from the tableviewer (e.g. huge 15,129 image matrix).
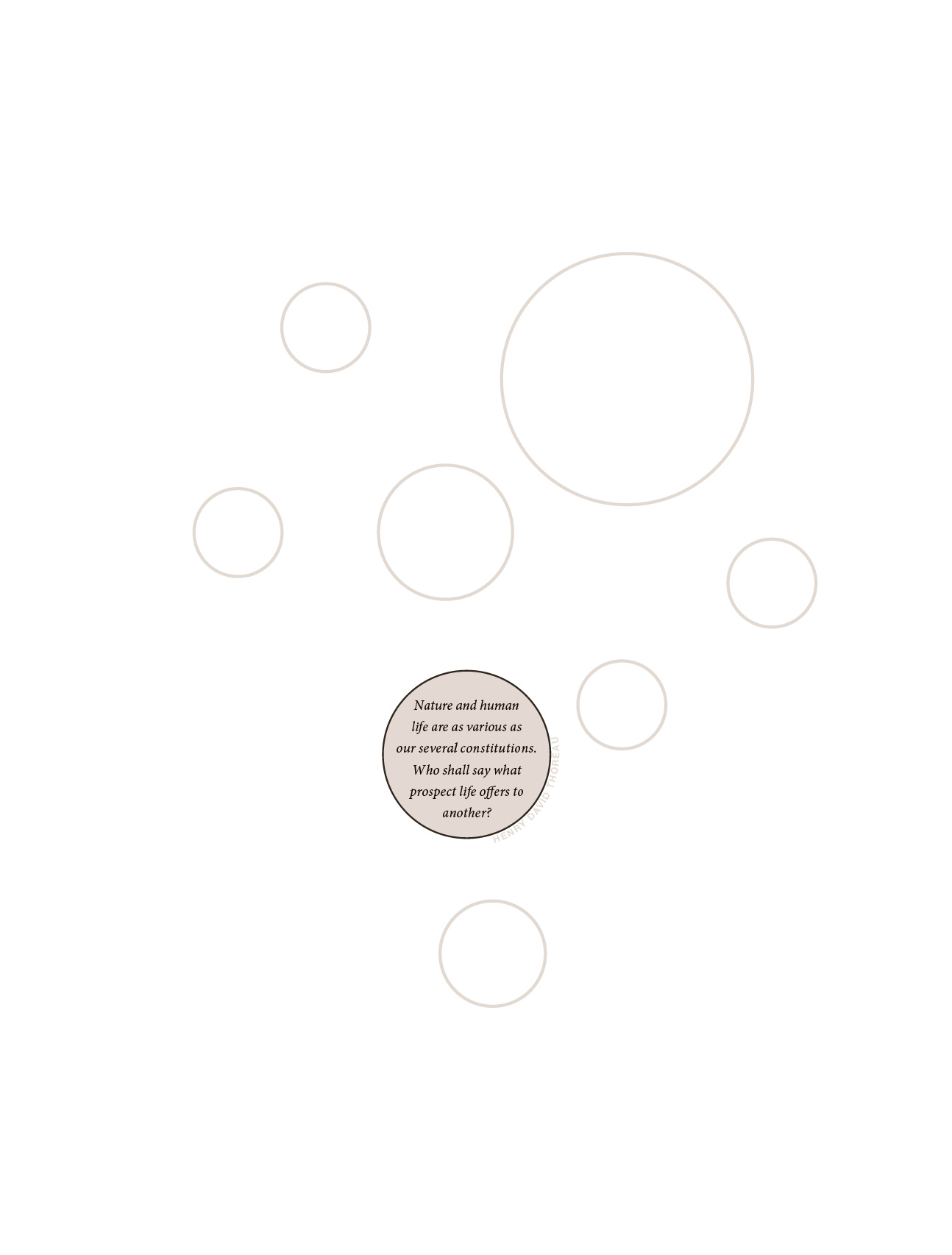
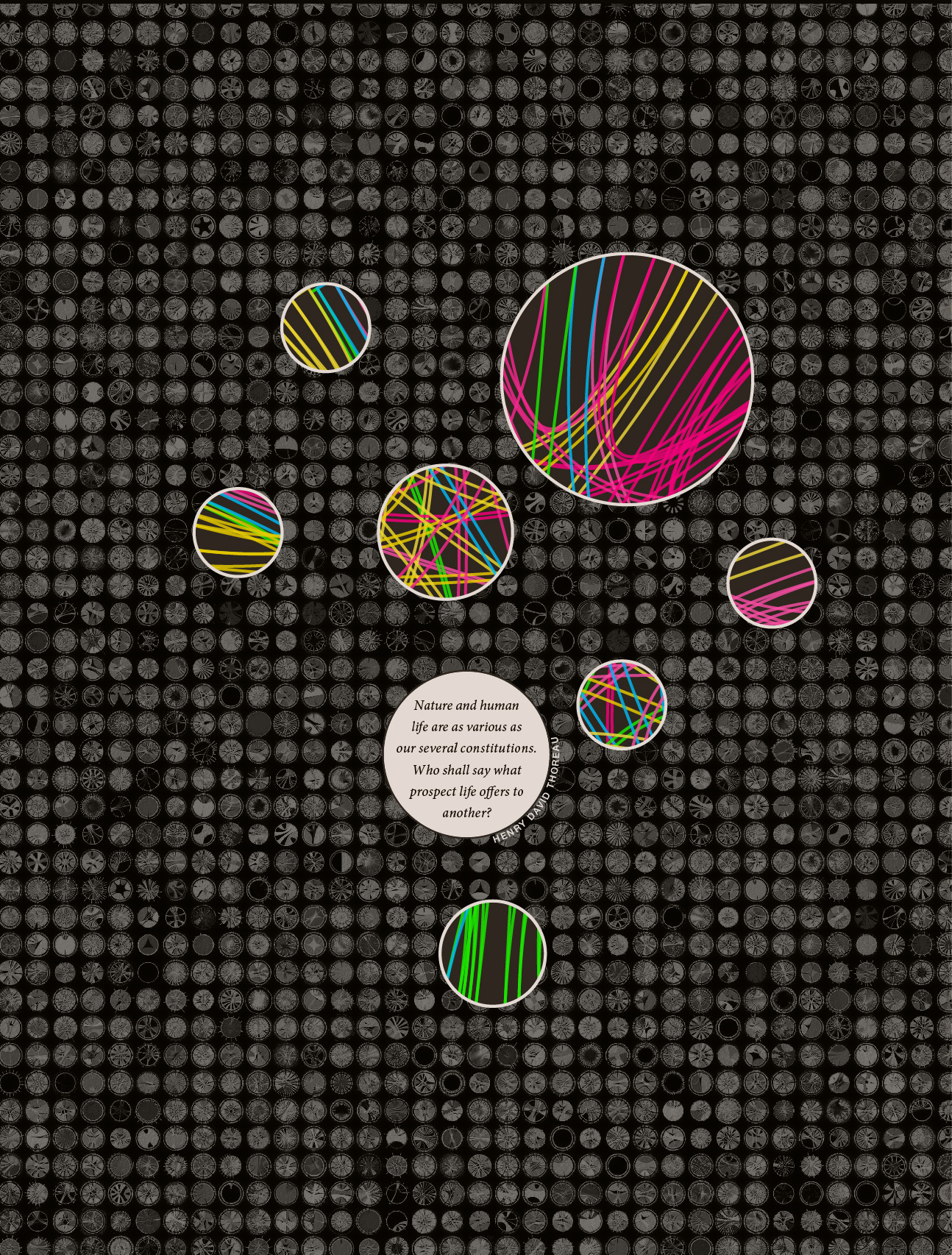
Rhiannon also wanted to include the quote by Henry David Thoreau, "Nature and human life are as various as our several constitutions. Who shall say what prospect life offers to another?" This reminded me of a similar but more tragic line from Shakespeare's Julius Caesar, "How many ages hence shall this our lofty scene be acted over in states unborn and accents yet unknown!"
In the early comps we played around with the idea of using non-genomics elements in the image, such as coins. We thought that we could use the variety of color and shape of the coins to communicate the idea of genetic diversity. However, after wrestling with how to do this effectively the concept was scrapped — the idea of using coins felt both arcane and arbitrary.
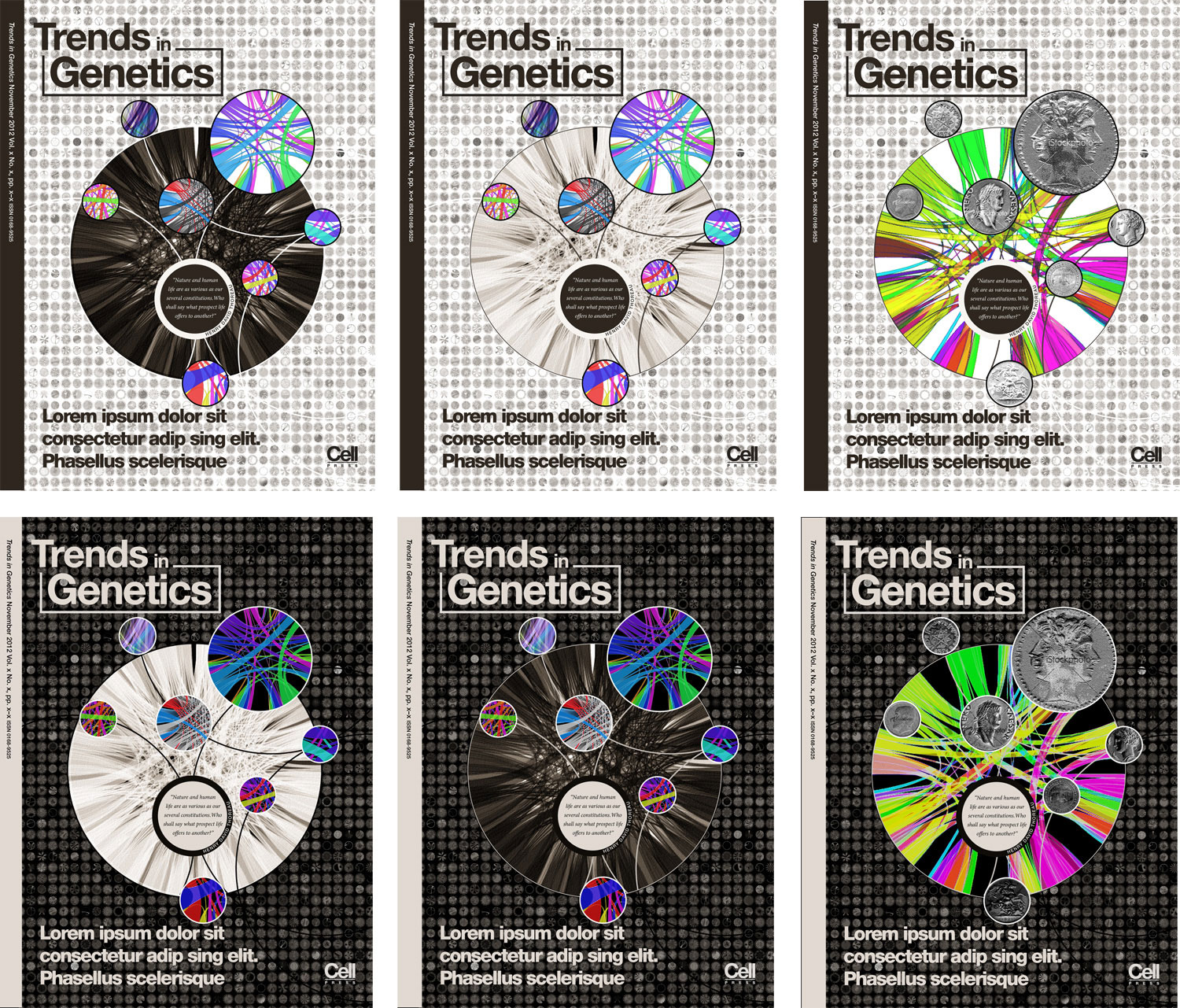
I decided to go with a warm brown color scheme. It's not a color I use a lot of, which makes me think that I should try to do more with it.
Deep brown provides great contrast for saturated colors, though I had to be careful not to make the image look too kitchy with an excess of colour variation. In some of the early comps shown above, two or more different color palettes were used (e.g. grey/red/blue and false color) and this lowered to overall visual cohesion of the image.
It's always a good idea to add variety to design. After all, without any variety we'd be left with a blank page. Ok, so variety is good, but too much variety is very bad, and can make you wish for that blank page again. Think about this: one kind of variety already provides variety! A variety of variety (I run the risk of recursing myself ad infinitum) can not only compete for attention but resonate destructively (that's design-speak for "turn into visual mush").
Everyone liked the combination of bright colors and dark background. This is an approach I favour too, which has worked well on other covers.

Briefly I experimented with various brush and pencil filters to give the image a more hand-drawn and organic look. Most of the illustrations I generate are very digital — blocks of solid colors and high-contrast shapes — and I thought a departure from this look could work in this case. However, like with the coins, this path didn't produce anything productive.



Nasa to send our human genome discs to the Moon
We'd like to say a ‘cosmic hello’: mathematics, culture, palaeontology, art and science, and ... human genomes.



Comparing classifier performance with baselines
All animals are equal, but some animals are more equal than others. —George Orwell
This month, we will illustrate the importance of establishing a baseline performance level.
Baselines are typically generated independently for each dataset using very simple models. Their role is to set the minimum level of acceptable performance and help with comparing relative improvements in performance of other models.
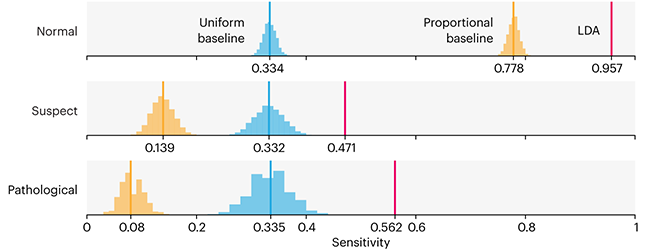
Unfortunately, baselines are often overlooked and, in the presence of a class imbalance5, must be established with care.
Megahed, F.M, Chen, Y-J., Jones-Farmer, A., Rigdon, S.E., Krzywinski, M. & Altman, N. (2024) Points of significance: Comparing classifier performance with baselines. Nat. Methods 20.
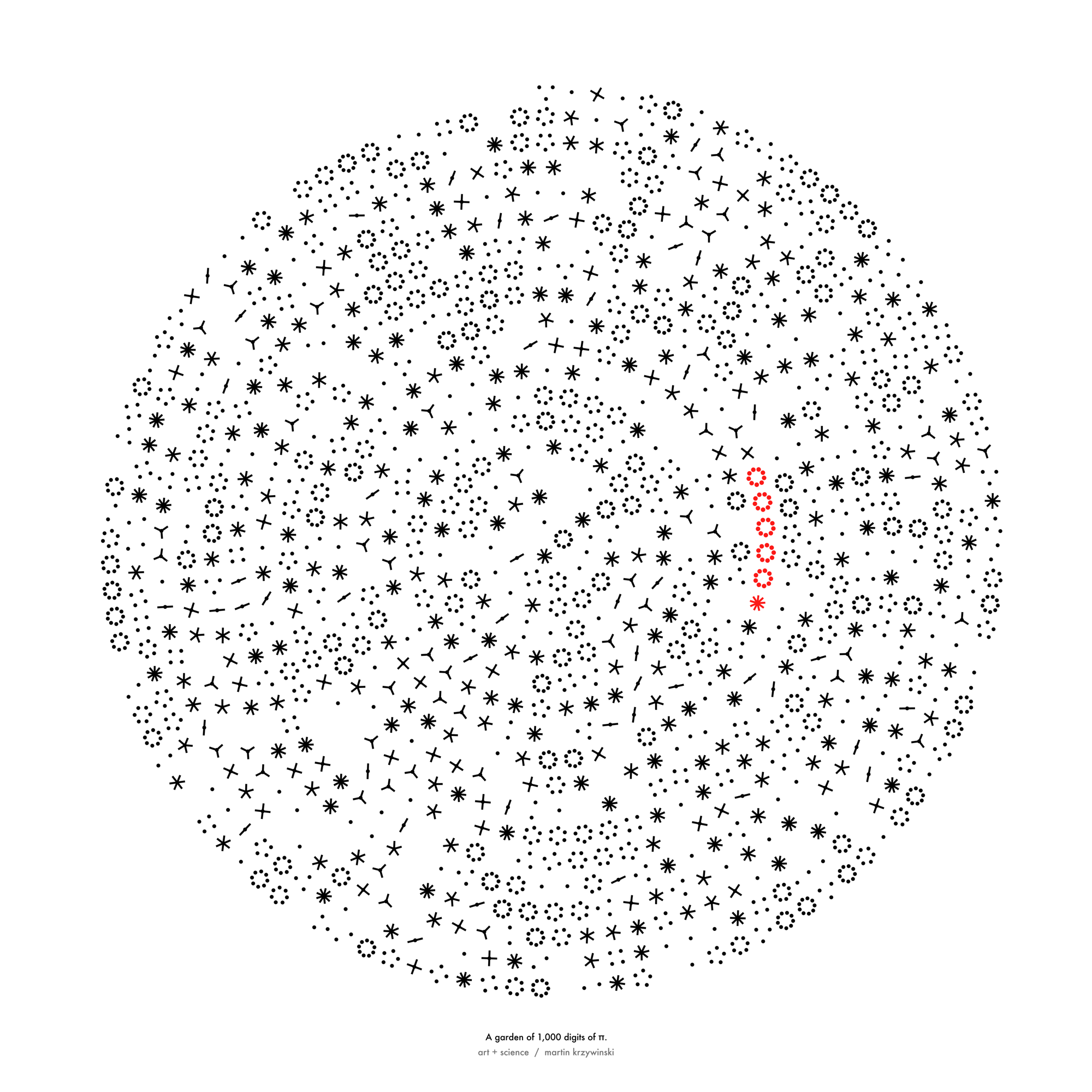
Happy 2024 π Day—
sunflowers ho!
Celebrate π Day (March 14th) and dig into the digit garden. Let's grow something.
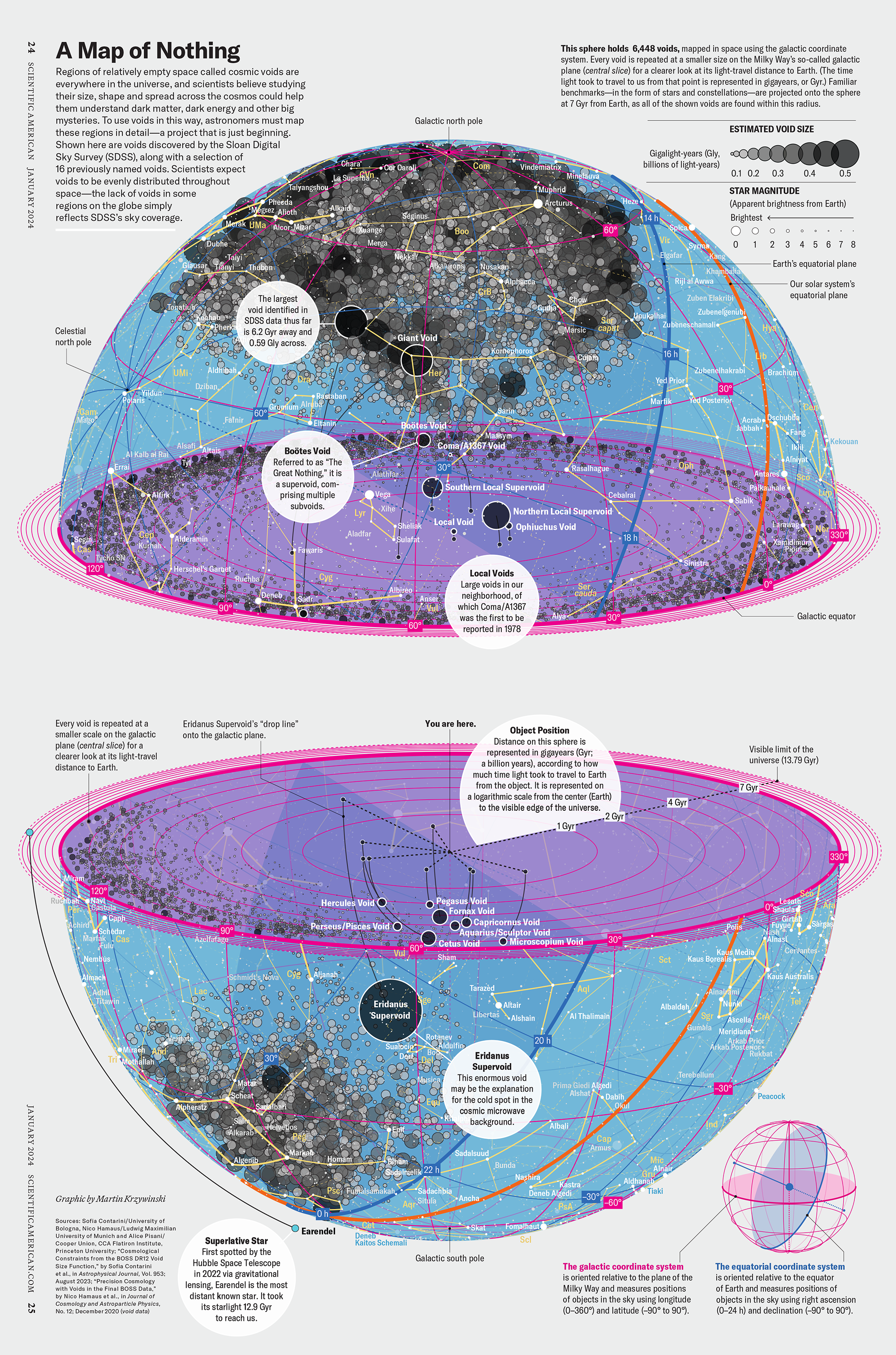
How Analyzing Cosmic Nothing Might Explain Everything
Huge empty areas of the universe called voids could help solve the greatest mysteries in the cosmos.
My graphic accompanying How Analyzing Cosmic Nothing Might Explain Everything in the January 2024 issue of Scientific American depicts the entire Universe in a two-page spread — full of nothing.
The graphic uses the latest data from SDSS 12 and is an update to my Superclusters and Voids poster.
Michael Lemonick (editor) explains on the graphic:
“Regions of relatively empty space called cosmic voids are everywhere in the universe, and scientists believe studying their size, shape and spread across the cosmos could help them understand dark matter, dark energy and other big mysteries.
To use voids in this way, astronomers must map these regions in detail—a project that is just beginning.
Shown here are voids discovered by the Sloan Digital Sky Survey (SDSS), along with a selection of 16 previously named voids. Scientists expect voids to be evenly distributed throughout space—the lack of voids in some regions on the globe simply reflects SDSS’s sky coverage.”
voids
Sofia Contarini, Alice Pisani, Nico Hamaus, Federico Marulli Lauro Moscardini & Marco Baldi (2023) Cosmological Constraints from the BOSS DR12 Void Size Function Astrophysical Journal 953:46.
Nico Hamaus, Alice Pisani, Jin-Ah Choi, Guilhem Lavaux, Benjamin D. Wandelt & Jochen Weller (2020) Journal of Cosmology and Astroparticle Physics 2020:023.
Sloan Digital Sky Survey Data Release 12
Alan MacRobert (Sky & Telescope), Paulina Rowicka/Martin Krzywinski (revisions & Microscopium)
Hoffleit & Warren Jr. (1991) The Bright Star Catalog, 5th Revised Edition (Preliminary Version).
H0 = 67.4 km/(Mpc·s), Ωm = 0.315, Ωv = 0.685. Planck collaboration Planck 2018 results. VI. Cosmological parameters (2018).
constellation figures
stars
cosmology
Error in predictor variables
It is the mark of an educated mind to rest satisfied with the degree of precision that the nature of the subject admits and not to seek exactness where only an approximation is possible. —Aristotle
In regression, the predictors are (typically) assumed to have known values that are measured without error.
Practically, however, predictors are often measured with error. This has a profound (but predictable) effect on the estimates of relationships among variables – the so-called “error in variables” problem.
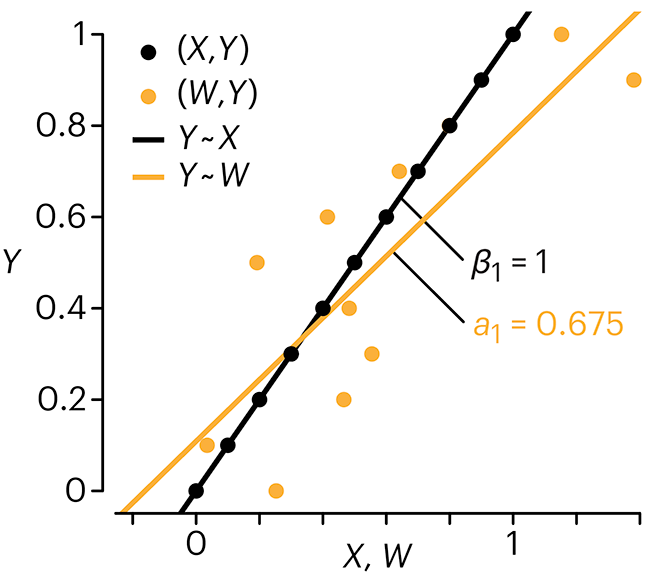
Error in measuring the predictors is often ignored. In this column, we discuss when ignoring this error is harmless and when it can lead to large bias that can leads us to miss important effects.
Altman, N. & Krzywinski, M. (2024) Points of significance: Error in predictor variables. Nat. Methods 20.
Background reading
Altman, N. & Krzywinski, M. (2015) Points of significance: Simple linear regression. Nat. Methods 12:999–1000.
Lever, J., Krzywinski, M. & Altman, N. (2016) Points of significance: Logistic regression. Nat. Methods 13:541–542 (2016).
Das, K., Krzywinski, M. & Altman, N. (2019) Points of significance: Quantile regression. Nat. Methods 16:451–452.
Convolutional neural networks
Nature uses only the longest threads to weave her patterns, so that each small piece of her fabric reveals the organization of the entire tapestry. – Richard Feynman
Following up on our Neural network primer column, this month we explore a different kind of network architecture: a convolutional network.
The convolutional network replaces the hidden layer of a fully connected network (FCN) with one or more filters (a kind of neuron that looks at the input within a narrow window).
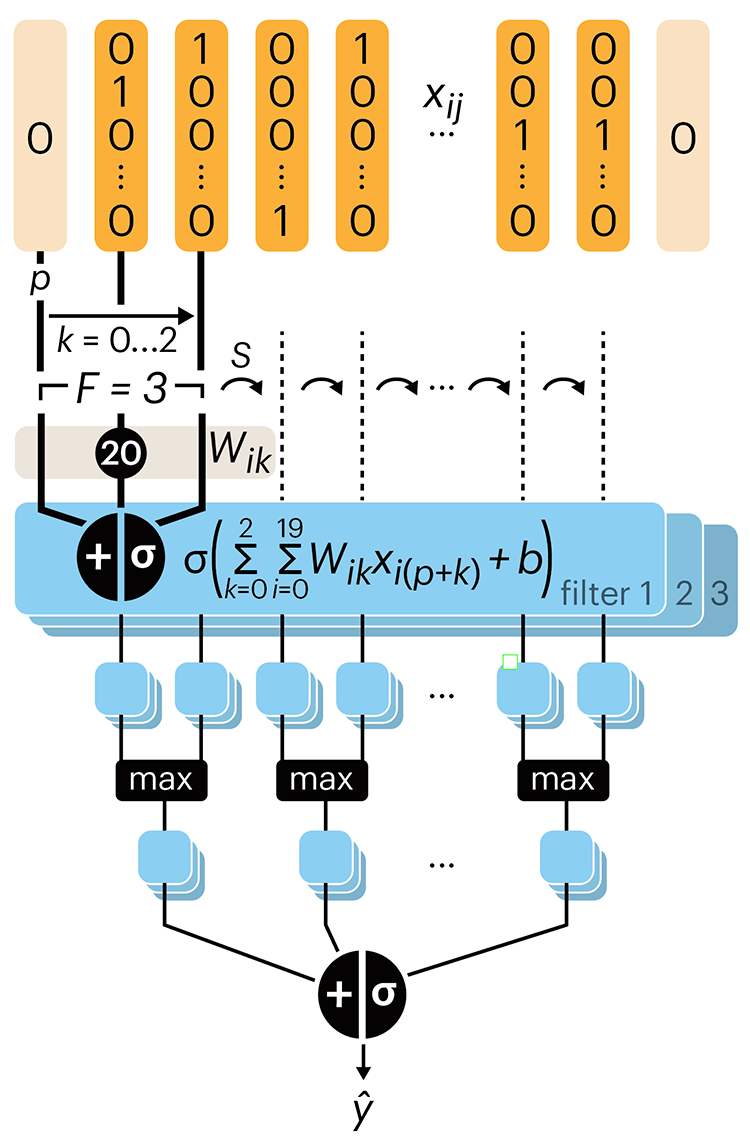
Even through convolutional networks have far fewer neurons that an FCN, they can perform substantially better for certain kinds of problems, such as sequence motif detection.
Derry, A., Krzywinski, M & Altman, N. (2023) Points of significance: Convolutional neural networks. Nature Methods 20:1269–1270.
Background reading
Derry, A., Krzywinski, M. & Altman, N. (2023) Points of significance: Neural network primer. Nature Methods 20:165–167.
Lever, J., Krzywinski, M. & Altman, N. (2016) Points of significance: Logistic regression. Nature Methods 13:541–542.