Art of the Personalized Oncogenomics Program
How often people speak of art and science as though they were two entirely different things, with no interconnection. An artist is emotional, they think, and uses only his intuition; he sees all at once and has no need of reason. A scientist is cold, they think, and uses only his reason; he argues carefully step by step, and needs no imagination. That is all wrong. The true artist is quite rational as well as imaginative and knows what he is doing; if he does not, his art suffers. The true scientist is quite imaginative as well as rational, and sometimes leaps to solutions where reason can follow only slowly; if he does not, his science suffers.
— Isaac Asimov, The Roving Mind (Ch 25)
I've prepared posters in three popular size formats: 11" × 14", 50 cm × 50 cm and 50 cm × 70 cm.
All artwork is available in PDF and PNG format. Click on the button on the top-right of the image to download these files. All files include 1/8" bleed. For printing, use the PDFs.
The PNG bitmap is provided for convenience and rastered at 600 dpi with 1/8" bleed (75 pixel margin on all sides). For example, the 11" × 14" bitmap has width 11.25 × 600 = 6,750 and height 14.25 × 600 = 8,550.
An explanation of how these images were generated, along with a printable legend, is available in the Methods section.
These posters are designed to fit a standard 11" × 14" frame.

These posters are fit to 50 cm × 70 cm and fit into inexpensive Strömby frames at IKEA.
The bigmap is 600 dpi (artboard 11,811 × 16,535 pixels) with 1/8" bleed (75 pixel margin on all sides).

These posters are fit to 50 cm × 50 cm and fit into inexpensive Strömby frames at IKEA.
You can print this poster to any square frame but keep in mind that if you shrink it down too much, the text may not be legible. At size, the text is 6.7 pt, which can be read comfortably. I would avoid printing the poster smaller than 30 cm × 30cm, which would have text of 4 pt in size.
The bigmap is 600 dpi (artboard 11,811 × 16,535 pixels) with 1/8" bleed (75 pixel margin on all sides).

This is the standard postcard size. The bitmap is 600 dpi (artboard 2,400 × 3,600 pixels) with 1/8" bleed (75 pixel margin on all sides).

Nasa to send our human genome discs to the Moon
We'd like to say a ‘cosmic hello’: mathematics, culture, palaeontology, art and science, and ... human genomes.



Comparing classifier performance with baselines
All animals are equal, but some animals are more equal than others. —George Orwell
This month, we will illustrate the importance of establishing a baseline performance level.
Baselines are typically generated independently for each dataset using very simple models. Their role is to set the minimum level of acceptable performance and help with comparing relative improvements in performance of other models.
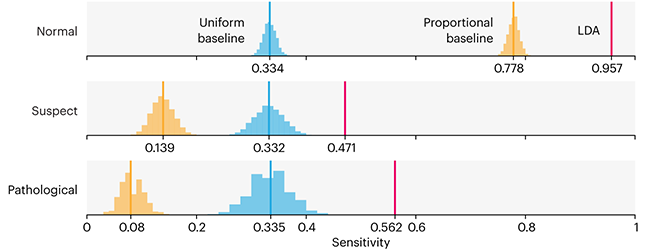
Unfortunately, baselines are often overlooked and, in the presence of a class imbalance5, must be established with care.
Megahed, F.M, Chen, Y-J., Jones-Farmer, A., Rigdon, S.E., Krzywinski, M. & Altman, N. (2024) Points of significance: Comparing classifier performance with baselines. Nat. Methods 20.
Happy 2024 π Day—
sunflowers ho!
Celebrate π Day (March 14th) and dig into the digit garden. Let's grow something.
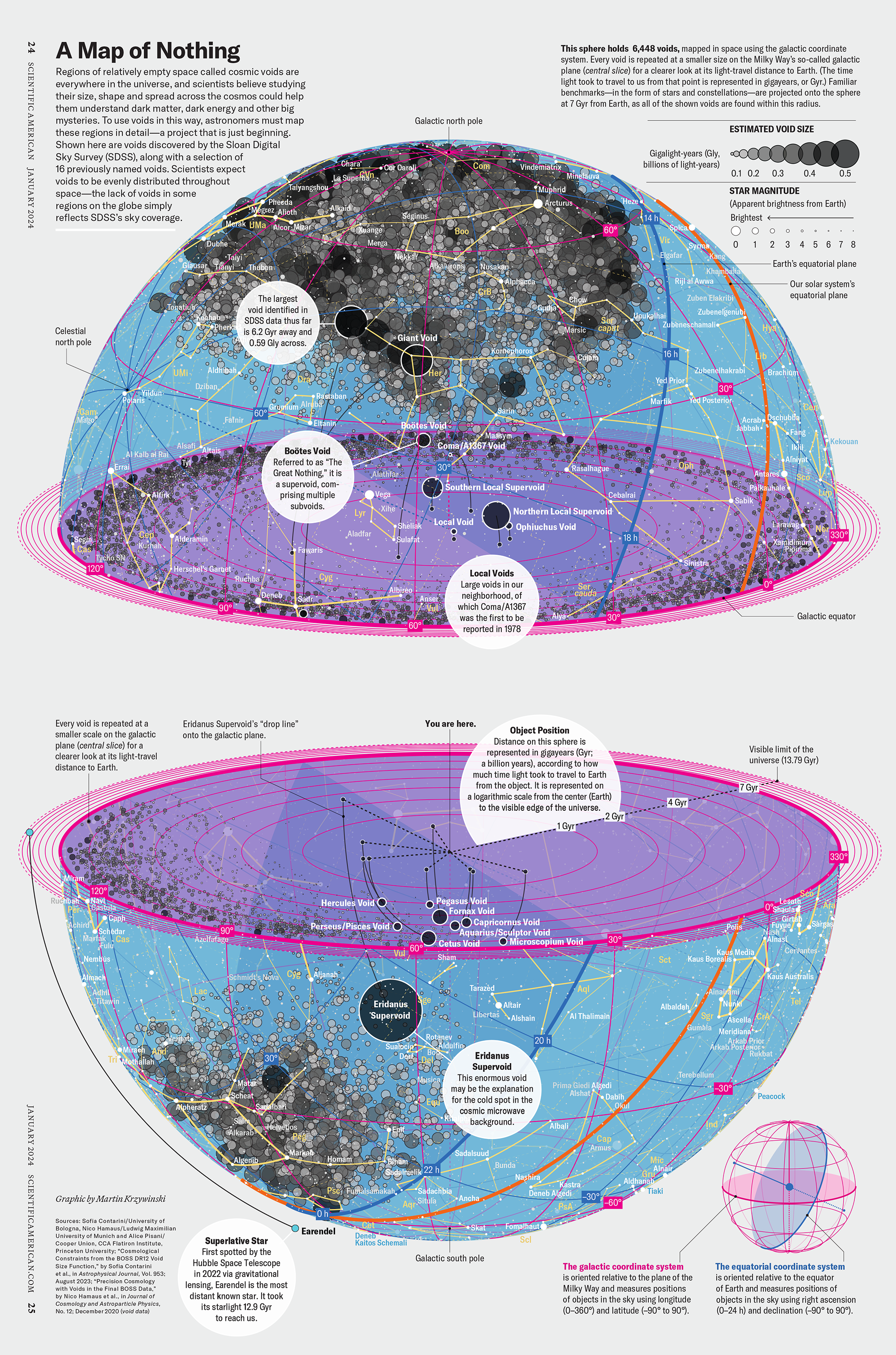
How Analyzing Cosmic Nothing Might Explain Everything
Huge empty areas of the universe called voids could help solve the greatest mysteries in the cosmos.
My graphic accompanying How Analyzing Cosmic Nothing Might Explain Everything in the January 2024 issue of Scientific American depicts the entire Universe in a two-page spread — full of nothing.
The graphic uses the latest data from SDSS 12 and is an update to my Superclusters and Voids poster.
Michael Lemonick (editor) explains on the graphic:
“Regions of relatively empty space called cosmic voids are everywhere in the universe, and scientists believe studying their size, shape and spread across the cosmos could help them understand dark matter, dark energy and other big mysteries.
To use voids in this way, astronomers must map these regions in detail—a project that is just beginning.
Shown here are voids discovered by the Sloan Digital Sky Survey (SDSS), along with a selection of 16 previously named voids. Scientists expect voids to be evenly distributed throughout space—the lack of voids in some regions on the globe simply reflects SDSS’s sky coverage.”
voids
Sofia Contarini, Alice Pisani, Nico Hamaus, Federico Marulli Lauro Moscardini & Marco Baldi (2023) Cosmological Constraints from the BOSS DR12 Void Size Function Astrophysical Journal 953:46.
Nico Hamaus, Alice Pisani, Jin-Ah Choi, Guilhem Lavaux, Benjamin D. Wandelt & Jochen Weller (2020) Journal of Cosmology and Astroparticle Physics 2020:023.
Sloan Digital Sky Survey Data Release 12
Alan MacRobert (Sky & Telescope), Paulina Rowicka/Martin Krzywinski (revisions & Microscopium)
Hoffleit & Warren Jr. (1991) The Bright Star Catalog, 5th Revised Edition (Preliminary Version).
H0 = 67.4 km/(Mpc·s), Ωm = 0.315, Ωv = 0.685. Planck collaboration Planck 2018 results. VI. Cosmological parameters (2018).
constellation figures
stars
cosmology
Error in predictor variables
It is the mark of an educated mind to rest satisfied with the degree of precision that the nature of the subject admits and not to seek exactness where only an approximation is possible. —Aristotle
In regression, the predictors are (typically) assumed to have known values that are measured without error.
Practically, however, predictors are often measured with error. This has a profound (but predictable) effect on the estimates of relationships among variables – the so-called “error in variables” problem.
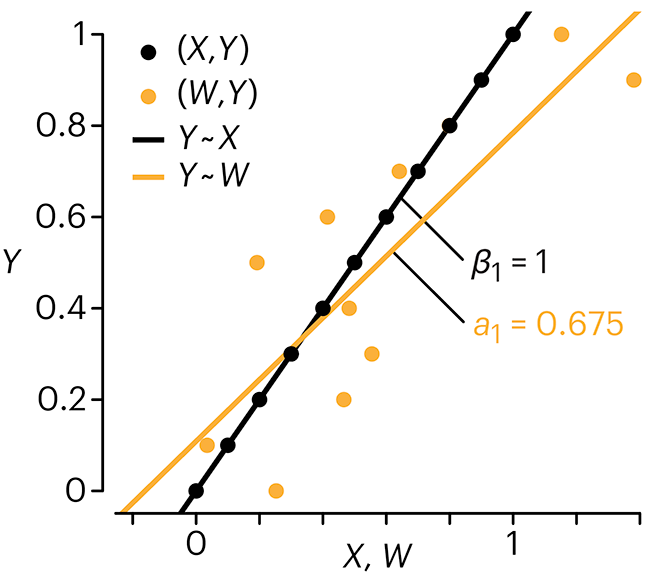
Error in measuring the predictors is often ignored. In this column, we discuss when ignoring this error is harmless and when it can lead to large bias that can leads us to miss important effects.
Altman, N. & Krzywinski, M. (2024) Points of significance: Error in predictor variables. Nat. Methods 20.
Background reading
Altman, N. & Krzywinski, M. (2015) Points of significance: Simple linear regression. Nat. Methods 12:999–1000.
Lever, J., Krzywinski, M. & Altman, N. (2016) Points of significance: Logistic regression. Nat. Methods 13:541–542 (2016).
Das, K., Krzywinski, M. & Altman, N. (2019) Points of significance: Quantile regression. Nat. Methods 16:451–452.
Convolutional neural networks
Nature uses only the longest threads to weave her patterns, so that each small piece of her fabric reveals the organization of the entire tapestry. – Richard Feynman
Following up on our Neural network primer column, this month we explore a different kind of network architecture: a convolutional network.
The convolutional network replaces the hidden layer of a fully connected network (FCN) with one or more filters (a kind of neuron that looks at the input within a narrow window).
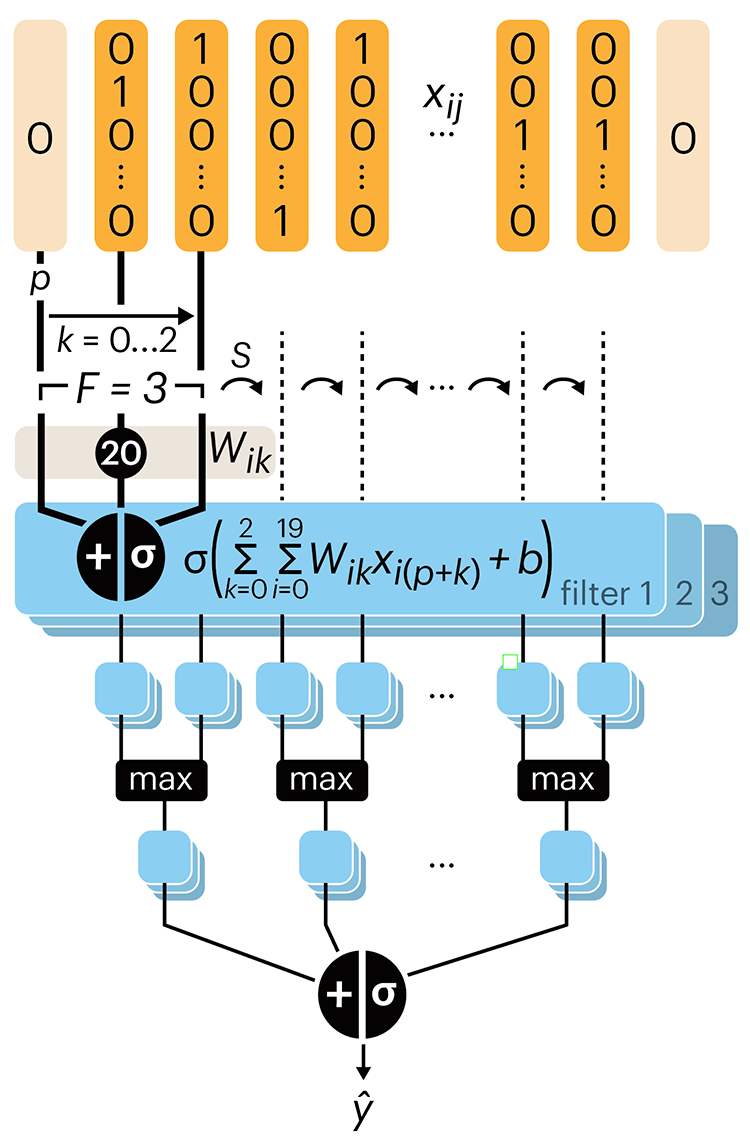
Even through convolutional networks have far fewer neurons that an FCN, they can perform substantially better for certain kinds of problems, such as sequence motif detection.
Derry, A., Krzywinski, M & Altman, N. (2023) Points of significance: Convolutional neural networks. Nature Methods 20:1269–1270.
Background reading
Derry, A., Krzywinski, M. & Altman, N. (2023) Points of significance: Neural network primer. Nature Methods 20:165–167.
Lever, J., Krzywinski, M. & Altman, N. (2016) Points of significance: Logistic regression. Nature Methods 13:541–542.