genomes + non-existence
Dark Matter of the Genome—sequences that are not there
Failing to fetch me at first keep encouraged,
Missing me one place search another,
I stop somewhere waiting for you.
—Walt Whitman, Song of Myself
One of the challenges in visualization is cueing what data is not being displayed, which is a common requirement when transitioning from overview to details, such as by zooming or filtering.
An interesting take on this is the enumeration and analysis of data that isn't merely not displayed but actually not there.
You can download the frequencies of all `k`-mers in the human genome for `k=1...12`.
I had an idea to try this and Michael Schatz pointed out an efficient algorithm [1] to find such missing sequences, or "nullomers" or, more generally, "unwords".
[1] Julia Herold, Stefan Kurtz and Robert Giegerich. Efficient computation of absent words in genomic sequences. BMC Bioinformatics (2008) 9:167
I wanted to create some phylogenetic trees based on these nullomers but discovered that this has also been done [2]. It turns out that the nullomers tell us something about the sequence without being visible. The genome's dark matter?
[2] Davide Vergni, Daniele Santoni. Nullomers and High Order Nullomers in Genomic Sequences. (2016) PLoS ONE 11(12): e0164540.
At some point I think I'll make some art around this concept.
sequences that don't exist
Because the human genome sequence is thankfully finite, there are some sequences that do not appear in it. These are (vaguely creepily) coined nullomers.
For example, the hg38 human reference assembly (Dec 2013), has 3,049,315,783 bases across its 455 assembled pieces, which includes the 24 chromosomes (e.g., chr1–chr22, chrX, chrY), mitochondrial chromosome (chrM), alternative and unanchored scaffolds (e.g. chr1_GL383520v2_alt and chr1_KI270706v1_random.fa). In this number I'm exluding all the padding in the sequence, which is indicated by the base "n".
Not surprisingly, all possible `k`-mers (sequences of length `k`) for `k=2` exist. There are 3,018,334,391 of them and they break down in frequency as follows
gc 130065644 4.31% ac 153830681 5.10% gt 154194068 5.11% cc 158048073 5.24% gg 159302235 5.28% tc 181782675 6.02% ga 182772932 6.06% ta 197567087 6.55% ct 213517855 7.07% ag 213785914 7.08% ca 221181041 7.33% tg 222266728 7.36% at 233904527 7.75% aa 296763858 9.83% tt 299351073 9.92% ---------- ----- 3018334391 100%
The least common 2-mer is "gc", which isn't surprising since the GC content of the human genome is about 41%, and both "g" and "c" appear in essentailly equal measure.
c 623727342 20.45% g 626335137 20.54% a 898285419 29.46% t 900967885 29.55% ---------- ------ 3049315783 100%
You can now ask "What is the smallest value of `k` for which we cannot find a `k`-mer in the human reference sequence?"
all `k`-mers up to `k=10` exist
It turns out that all possible sequences up to length 10 exist in the reference.
Depending on your familiarity with the genome and depth of intuition about statistics, this might be not at all surprising, mildly suprising or just plain confusing.
For example, if we consider `k=8`, there are `4^8 = 65536` such sequences. Alphabetically,
aaaaaaaa aaaaaaac aaaaaaag aaaaaaat aaaaaaca ... ttttttgt ttttttta tttttttc tttttttg tttttttt
All these exist. The 8-mer "cgtacgct" is the most rare and "tttttttt" the most common.
cgtacgcg 304 0.00000997% cgcgtacg 315 0.00001033% cgtcgacg 322 0.00001056% tcgacgcg 333 0.00001092% cgatcgac 356 0.00001167% ... tgtgtgtg 899632 0.02950286% tatatata 925963 0.03036637% atatatat 967263 0.03172078% aaaaaaaa 4743237 0.15555144% tttttttt 4773492 0.15654364%
If we repeat this all the way up to `k=10`, we find that all sequences exist. For example, there are `4^{10}=1,048,576` 10-mers and they also all exist.
cgacgatcga 2 0.00000007% tcgcgacgta 2 0.00000007% cgtaacgcgc 2 0.00000007% tcgacgtacg 2 0.00000007% tattcgcgcg 2 0.00000007% ... cacacacaca 602118 0.01974610% gtgtgtgtgt 604553 0.01982595% tgtgtgtgtg 608965 0.01997064% aaaaaaaaaa 3085421 0.10118453% tttttttttt 3105565 0.10184514%
The least common 10-mer is "cgacgatcga", which appears only twice. In fact, the 5 10-mers shown above are the only ones that appear only twice. No 10-mer appears once.
Once we look at 11-mers, we finally see a sequence that doesn't appear. There are `4^{11} = 4,194,304` 11-mers of which 2,728 (0.07%) appear once and 4,705 (0.11%) appear twice. The most common ones are
ctgggattaca 409261 0.01342148% tgtaatcccag 409332 0.01342380% ctgtaatccca 420484 0.01378953% tgggattacag 420548 0.01379163% cacacacacac 512570 0.01680943% gtgtgtgtgtg 517147 0.01695953% acacacacaca 549346 0.01801548% tgtgtgtgtgt 555966 0.01823258% aaaaaaaaaaa 2563770 0.08407734% ttttttttttt 2580640 0.08463058%
once you take out what's not there, you're left with what's there
Interestingly (maybe) 941 11-mers don't appear in the genome. I've listed them alphabetically below. The first one is the most interesting one, as it is the first alphabetically ordered sequence that does not appear in the genome: aaccgacgcgt.
If you look at the statistics of all the bases of these missing 11-mers, you find that they're the inverse of what is there.
# 11-kmer nullomers # bases in hg38 c 3166 30.59% 623727342 20.45% g 3182 30.74% 626335137 20.54% a 1986 19.19% 898285419 29.46% t 2017 19.49% 900967885 29.55%
For example, since 41% of the bases in the genome are "g" or "c" we find `0.305 \approx (1-0.41)/2` of the missing bases to be "g" or "c" and similarly `0.19 \approx (1-0.59)/2` to be "a" or "t".
# all 11-mers sorted alphabetically that do not appear
# in the human genome reference sequence (hg38 Dec 2013)
aaccgacgcgt aaccgcgcgta aaccgcgtacg aaccggttcgc aacgcgatacg aacgcgatcgg aacgcgcgaat aacgcggtcga aacgcgtagcg aacgcgtatcg aacggtcgtcg aacgttgcgcg aagcgtacgcg aagtcgcgcga aatacgcgcga aatacgtcgcg aatcgcgtacg aatcgcgtcga aatcgcgttcg aatcggttgcg aatcgtcgacg aatgtcgcgcg aattcgcgacg aattcgcgcga aattgtcgcga acacgcgtcga acccgtcgcga accgatacgcg accgatcgacg accgatcgtcg accgccgatcg accgcgacgaa accgcgcgatt accgcgtacgt accggtcggac accgtcgatcg accgtcgcgat accgtcgcgta accgttcgtcg acgaaacgtcg acgaatcgacg acgaccgcgta acgaccgttcg acgacgacgta acgacgccgta acgacgcgcta acgacgcgtat acgacggtacg acgatacgcga acgatacgcgg acgatacggcg acgatcggcga acgatcgtcgg acgatcgtgcg acgattcggcg acgcaacgtcg acgccggtcga acgcgacgatt acgcgacgcta acgcgacggta acgcgacgtaa acgcgataacg acgcgatacgt acgcgcgatac acgcgcgatat acgcgcgatta acgcggcgtaa acgcggtacga acgcggtacgt acgcgtaacga acgcgtaacgt acgcgtaccgg acgcgtacgat acgcgtcgtac acgctagtcga acgctcgaacg acggcgactaa acgggtcgacg acggtacgccg acggtacgtcg acggtcgagcg acggtcgcgta acgtaacgcgc acgtaccgtcg acgtacgaacg acgtacgatcg acgtagcgcgt acgtatcgccg acgtcgaccga acgtcgacgct acgtcgacgta acgtcgagtcg acgtcgcgata acgtcgcgcgt acgtcgcgtac acgtcggtcga acgtcgtacgc acgttaacgcg actaatcgcga actacgcgtcg acttcgcgcgt agcgtcgtacg agcgttcgacg agtacgtcgcg agtatcgcgac agtcgatcgcg agtcggtcgat agttacgcgcg ataccgcgtcg atacgaacgcg atacgacgcga atacgcgatcg atacgcgccga atacgcgcgac atacgcgcgag atacgcgttcg atacggcgcgt atacttcgccg atacttcgcgc atagactcgcg atatcgacgcg atatcgcgcgg atatcgcgcgt atatcgtcgcg atccgtcgcga atcgacgtgcg atcgcgaaacg atcgcgacgta atcgcgattcg atcgcgcgaag atcgcgcgtat atcgcgcgtcg atcgcggtcgt atcgcgtaacg atcgcgtaccg atcgcgtacgt atcgcgttccg atcggcgcgta atcgggtcgac atcggtacgcg atcggttgcga atcgtaccgcg atcgtccgacg atcgtcgaacg atcgtcgacga atcgtcgacgc atcgtcgatcg atcgtcgcgta atcgtctcgcg atgtatcgcgc atgtcgcgcga atgtcgtcgcg attcgacggcg attcgcgatcg attcgcgcgat attcgcggcgc attgtacgcgg atttcgacgcg atttcgcgacg atttcgcgcga atttcgtcgcg caacgcgctac cacgtcgcgaa cagtccgatcg catacgcgtcg catatcgcgcg ccgaatacgcg ccgaccgtacg ccgacgatcga ccgacgatcgt ccgacgcgata ccgacgcgatt ccgacgtaacg ccgacgtacga ccgatacgcga ccgatacgtcg ccgatagtcgg ccgatcgtccg ccgattacgcg ccgcgaatcga ccgcgacgtaa ccgcgataacg ccgcgatacga ccgcgcgatat ccgcgtaatcg ccgcgtcgaaa ccgcttaagcg ccggcgtaacg ccgtataacgg ccgtcgaaacg ccgtcgaacgc ccgtcgaccgg ccgtcgatcga ccgtcgccgaa ccgtcgcgtag ccgtcgcgtat ccgttacgtcg ccgttcgcacg ccgttgcgtcg cgaacggtcgc cgaacggtcgt cgaacggttcg cgaacgtacga cgaactaacgg cgaatacgcgc cgaataggcgg cgaatcgacga cgaatcgacgg cgaatcgagcg cgaatcgcgta cgaatcgtcga cgacagtacgg cgacatacgcg cgaccgactat cgaccgatacg cgaccgctata cgaccgtcgac cgaccgtcgat cgaccgttaac cgaccgttacg cgacgaacgag cgacgaacggt cgacgaccgta cgacgacgtat cgacgatacgg cgacgatcgaa cgacgatcgat cgacgatcggc cgacgattcgc cgacgcaacga cgacgcgacaa cgacgcgacat cgacgcgataa cgacgcgatac cgacgcgtaaa cgacgcgtata cgacgcgtcaa cgacgcgttac cgacgcgttta cgacgctatcg cgacggacgta cgacggatacg cgacggcgtat cgacgtaacgc cgacgtaacgg cgacgtaccga cgacgtaccgt cgacgtacgac cgacgtacgat cgacgtacggg cgacgtactcg cgacgtatcgg cgactacgcga cgactagtccg cgactatcgcg cgagcgtatcg cgagtcgaccg cgataccgcgt cgataccgtcg cgatacgaccg cgatacgcgag cgatacgcgat cgatacgcgga cgatacggcgt cgatacgttcg cgatagacggt cgatagcgcgt cgatagtcgga cgatagtcggt cgatatacgcg cgatatgcgcg cgatccgatac cgatccgtacg cgatcgaccga cgatcgaccgt cgatcgacgtc cgatcgcgaac cgatcggcgta cgatcggtcgt cgatcgtacgc cgatcgtacgg cgatcgtcggg cgatcgtcgta cgatcgtgcga cgatgcgcgac cgattacgcga cgattacgcgc cgattacgcgt cgattatcgcg cgattcgacgt cgattcggcga cgattgaccgc cgcaacggtac cgcaatagtcg cgcatatcgcg cgcattcgtcg cgccgaaacga cgccgatacga cgccgatcgaa cgccgatcgta cgccgtaatcg cgccgtacgta cgcgaaacgat cgcgaacgatt cgcgaacgtta cgcgaatcgaa cgcgaattcgt cgcgaccgaat cgcgaccgtaa cgcgacgaact cgcgacgatta cgcgacgcata cgcgacgctaa cgcgacgctat cgcgacggtta cgcgacgtaac cgcgacgttaa cgcgacttacg cgcgagcgata cgcgagcgtag cgcgataacga cgcgataacgc cgcgataattg cgcgatacacg cgcgatcaacg cgcgatcggta cgcgattcgat cgcgattgtcg cgcgcacgata cgcgcataaga cgcgcataata cgcgcgatatg cgcgcgatatt cgcgcgtatta cgcgcgttata cgcgctatacg cgcgctatccg cgcggtacgaa cgcggtacgta cgcggtatacg cgcggtcgatt cgcggttcgtt cgcgtaacgcg cgcgtaatacg cgcgtaatcga cgcgtaatcgt cgcgtaccgga cgcgtaccgtt cgcgtatcgcg cgcgtatcggt cgcgtatcgtt cgcgtattcgg cgcgtcaaacg cgcgtcgagac cgcgtcgcaat cgcgtcggatg cgcgtcgtact cgcgtcgttat cgcgttacgcg cgcgttatacg cgcgttattcg cgcgtttcgta cgctaacgtcg cgctcgacgaa cgctcgacgta cgctcgcgtat cgctcgtaacg cgctcgtacga cgctcgtatcg cgcttacgcga cggaccatacg cggactatcga cggagcgtacg cggatcgacga cggcgaacgta cggcgatctaa cggcgcgatag cggcgtaccga cggcgttaacg cggcttacgcg cggtaatcgcg cggtacgatcg cggtacgccga cggtatacggg cggtccgcata cggtcgaccgt cggtcgataac cggtcgattcg cggtcgcacga cggtcgcgtaa cggtcggtacg cggtcgtacga cggttacgtcg cggttcgatcg cggttcgtacg cgtaacgagcg cgtaacgccgt cgtaacgcgca cgtaacgcgct cgtaacgcgtt cgtaacgctcg cgtaacgtccg cgtaacgttcg cgtaccggtct cgtacgaacgc cgtacgaatcg cgtacgaccgt cgtacgacgaa cgtacgacgct cgtacgatacg cgtacgatcgg cgtacgatcgt cgtacgatgcg cgtacgccgtt cgtacgcgaat cgtacgcgact cgtacgcgata cgtacgcgatc cgtacggacgc cgtacggcggt cgtacggtcga cgtacggtcgt cgtacgtcgcg cgtacgtcggc cgtacgtgacg cgtatacgacg cgtatacgcga cgtatagcgcg cgtatatcggc cgtatcgcgtc cgtatcggtcg cgtatgtcgcg cgtattacgcg cgtattcgacg cgtattcgcgc cgtattgcgcg cgtcaatcgcg cgtcacgcgta cgtccgatcgt cgtccgtcgaa cgtcgaacgat cgtcgaattcg cgtcgaccggt cgtcgaccgtc cgtcgacgatc cgtcgacgcgt cgtcgacgctt cgtcgacgtac cgtcgactacg cgtcgactatc cgtcgagacgt cgtcgagcatc cgtcgataggc cgtcgattcga cgtcgcgaagc cgtcgcgacta cgtcgcgatac cgtcgcgatag cgtcgcgtagg cgtcgcgtatg cgtcgcgtatt cgtcgctcgaa cgtcggaatcg cgtcggtacga cgtcggtcgac cgtcggtcgat cgtcggttacg cgtcgtaccgg cgtcgttatac cgtcgttcgac cgtctcgtacg cgtgcgaacta cgtgtcgaacg cgtgtcgacga cgttaaacgcg cgttacgacga cgttacgcgtc cgttacggcgt cgttacggtcg cgttacgtcgt cgttagtcgcg cgttccgtcga cgttcgacgaa cgttcgacgcg cgttcgacggc cgttcgacggg cgttcgatcgt cgttcgcgaaa cgttcgcggaa cgttcgcggta cgttcgcgtat cgttcgctagg cgttgcgatcg cgttgcgtcgt cgtttacgcga cgtttcgaccg cgtttcgcgaa cgtttcgtcga ctaccgccgta ctacgaacggt ctacgcgcgaa ctacgcgcgac ctacgcgcgta ctacgcgtcga ctacgtcgacg ctactcgatcg ctagacgtacg ctataccgcgc ctatacgtccg ctatcgcgtcg ctatcgtcgcg ctattcgcgcg ctcgaccgtcg ctcgacgcgta ctcgacgtacg ctcgatcgacg ctcgatcgtcg ctcgcgacgta ctcgcgcggta ctcgtcgagta ctcgttcgtcg cttcgcgaacg gaccgatacgc gacgacgatcg gacgcgataat gacgcgattcg gacgcgcatag gacgcgcgtat gacgcgtaacg gacgtaacgcg gacgtacggcg gacgtacgtcg gacgtcgaacg gatacgtcgcg gatacttcgcg gatagcgcgtt gatagtcgacg gatcgcgaacg gatcgcgcgta gatcggtccga gatcggtcgta gatcgtcggtc gcccgttcgta gccgatcgttg gcgaacgcgta gcgaattacgc gcgacgatacg gcgacgatcga gcgacgcgata gcgacgcgtta gcgacgtaacg gcgattgacga gcgcatatcgt gcgccggtata gcgcgaatcga gcgcgacgata gcgcgacgtta gcgcgtaccga gcgcgtacgat gcgcgttcgat gcggtacgcgt gcggtcgacga gcggtcgtacg gcgtaacgcgc gcgtacaacga gcgtacgtcgc gcgtcgacaat gcgtcgattcg gcgtcgattgt gcgtcgtacgc gcgttacgcgt gcgttcgacgg gcgttcgcgta gctacgaaccg gctatacgcgt gctatcgcgcg gctcgtatcgt ggcgcgctata ggcgtcgatcg ggcgtcgcaat ggtacgcgacc ggtacgcgtaa ggtataccgcg ggtcgacgcga ggtcgattcgc ggtcgattgcg ggttccgtacg gtaatcgacga gtaccggcgta gtacgaggtcg gtacgccggtt gtacgcgaccg gtacgcgagta gtacgcgcgta gtacgcgcgtt gtacgctcgac gtacggcgatc gtacggcgtac gtacggtcgcg gtacgttcgcg gtagcggtacg gtataacgcgg gtatagcgacg gtatccgatcg gtatcgaaccg gtatcgacgcg gtatcgcgcga gtatcgcgtcg gtatcgtcgcg gtatcgttgcg gtatgccgcga gtatgcgaacg gtattatcgcg gtattcgacgc gtccgacgcga gtccgacgtcg gtccgagcgta gtccgatcgcg gtccgttacgc gtcgaaccgac gtcgaacgacg gtcgaacgcga gtcgacgacta gtcgacgatcg gtcgacgcgaa gtcgacgcgac gtcgacgtacg gtcgatcggta gtcgattcgag gtcgcaattcg gtcgcacgcga gtcgcccgata gtcgcgacaat gtcgcgacgta gtcgcgcataa gtcgcgccgta gtcgcgcgtaa gtcgcggataa gtcgcgtcgca gtcgcgttacg gtcgctatcgt gtcgtaacgcg gtcgtacgcga gtcgttacgcg gttatgcgcga gttattcgcgc gttcgaacgcg gttcgatcgga gttcgtacgcg gttcgtagcga taaccgtcgac taacgcgatcg taacgcgccga taacgcgcgat taacgcgtcga taacgtcgcga taacgtcgcgc taagtcgcgcg taatcgcgtcg taatcgtcgac taattcgacgc taattcgcgcg tacccgcggtt taccgcgcgat taccggtcgta taccgtacgcg taccgttcgcg tacgacgcgat tacgacgtacg tacgagctcgt tacgatcgacg tacgatcgtcg tacgcacgcga tacgccgacgc tacgcgacacg tacgcgaccga tacgcgacgag tacgcgacgca tacgcgatcga tacgcgatcgc tacgcgatgcg tacgcgattcg tacgcgcgaac tacgcgcgaca tacgcgcgagc tacgcgcgata tacgcgcgatt tacgcgcggtc tacgcgcgtaa tacgcggtacg tacgcgtaacc tacgcgtacga tacgcgtatcg tacgcgtcacg tacgcgtcgac tacgctacggc tacgctcggac tacggcgtacg tacgggcgacg tacgggcgtcg tacggtcgcga tacgtacgcga tacgtccgacg tacgtccgtcg tacgtcgagcg tacgtcgatcg tacgtcgcgca tacgtcgcgct tacgtcgcgta tacgtcgctcg tacgtcggtcg tacgtcgtacg tacgtcgtcgc tacgttacgcg tacgttcgacg tactacgcgcg tactatcgacg tactcgcgacg tagaccgacgc tagccggtacg tagccgttcga tagcgcgaatc tagcgcgacgt tagcgcgtcga tagcgtaccga tagctcgacga taggcgaaccg taggcgcgtaa tagtcgacgca tagttacgcgc tatacgcgaac tatacgcgcga tatacgcgtcg tatatgtcgcg tatccgatcgc tatccgcgcgt tatccggatcg tatccgtcgca tatcgccgact tatcgcgcgca tatcgcgcgct tatcgcggtcg tatcgcgtcga tatcgcgtcgt tatcgcgttcg tatcgctcgac tatcggcgatc tatcgtcgacg tatcgtcgccg tatcgttcgcg tatgcgaccgc tatgcgcgacg tatgcgcgcga tatgcgtcgcg tatggcgcgcc tatgtcgacgc tatgtcgcgat tatgtttcgcg tattacgcgcg tattatgcgcg tattcgacgcg tattcgcgacg tattcgcgcga tattcgcgcgg tattcgcggcg tattcgcgtcg tattcggcgcg tcatatcgcgc tccgatagtcg tccgcgactta tccgcggtacg tccgtaacgcg tccgtacgcga tccgtacgcgg tccgtcgaccg tccgtcgatcg tcgaacgatcg tcgaatacgcg tcgaccgcgta tcgaccgtcga tcgaccgtcgg tcgaccgttcg tcgacgaacga tcgacgaccga tcgacgaccgt tcgacgagtcg tcgacgatcgc tcgacgcgata tcgacgcggtt tcgacgcgtag tcgacggtacg tcgacggtatg tcgacgtaccg tcgacgtacga tcgacgtacgg tcgacgttacg tcgactaagcg tcgactagcgg tcgactcgacg tcgagccgacg tcgagcgcgta tcgagcgtacg tcgatacgccg tcgatacgcgg tcgatcgacgt tcgatcggata tcgatcgtacg tcgatcgtcgg tcgatgcgtcg tcgattacgcg tcgattagcgc tcgatttcgcg tcgcaatcggc tcgcacgatcg tcgcacgcgat tcgccgaatcg tcgccgtacgg tcgcgaaacgt tcgcgaaccga tcgcgaacgtt tcgcgaatacg tcgcgacccgt tcgcgaccgta tcgcgacgcaa tcgcgacgcga tcgcgacgtaa tcgcgacgtag tcgcgactcgt tcgcgagcgta tcgcgattcga tcgcgccgata tcgcgcgaacg tcgcgcgaata tcgcgcgacat tcgcgcgacta tcgcgcgagta tcgcgcggcta tcgcgcgtcaa tcgcgcgttga tcgcggatacg tcgcgtaatcg tcgcgtatacg tcgcgtatcgc tcgcgtcaacg tcgcgtccgat tcgcgtcgaac tcgcgtcggta tcgcgtcgtta tcgcgttaacg tcgcgtttcga tcgctcgcgat tcgctcgtcga tcggacgtacg tcggacgtagc tcggcgatacg tcggcgatata tcgggctatcg tcggtacgcgc tcggtacgcta tcggtcgacga tcggtcgtacg tcgtaacgcgc tcgtaatacgg tcgtacgaccg tcgtacgccga tcgtacggccg tcgtatcgccg tcgtatcgtcg tcgtccgatcg tcgtcgaaccg tcgtcgaatcg tcgtcgacgat tcgtcgattcg tcgtcgcgata tcgtcgcgcaa tcgtcgcgtac tcgtcggtacg tcgtcggtcga tcgtctcgcgt tcgttacgcgg tcgttcgaccg tcgttcgagcg tcgttcgcgta tcgttcgtacg tcgtttcgacg tctacgcgtcg tgacgaacgcg tgatcgcgacg tgatcgcgtag tgcgaacgacg tgcgcggcgta tgcgtaacgcg tgcgtcgtacg tgtacgcgacg tgtcgacgcga tgtcgacgcgt tgtcgcgcgat tgtcgcgcgta tgtcgcgtcgt ttaaccgtcga ttaacgcgcga ttaacgtcgcg ttaccgcgacg ttacgacgcgt ttacgcgacgc ttacgcgatcg ttacgcgcgcc ttacgcgcgga ttacgcggaca ttacgcgtacc ttacgtcgcga ttagcgcgtca ttaggtcgcgc ttagtacgcgc ttagtcgctcg ttatcgcgccg ttatcgcgcgc ttattcgcgcg ttcgacgaacg ttcgacgcgac ttcgacgcgag ttcgacgcgta ttcgacggacg ttcgagcgacg ttcgatccgtt ttcgccgatcg ttcgcgagacg ttcgcgattcg ttcgcgcatag ttcgcgcgaat ttcgcgcgata ttcgcgtacgg ttgattacgcg ttgcgcgatag ttgtcgacgcg tttagtcgcgc tttcgacgcaa tttcgacgcgt tttcgcgtacg tttcgtcgcgc tttcgtcgcgg
Nasa to send our human genome discs to the Moon
We'd like to say a ‘cosmic hello’: mathematics, culture, palaeontology, art and science, and ... human genomes.



Comparing classifier performance with baselines
All animals are equal, but some animals are more equal than others. —George Orwell
This month, we will illustrate the importance of establishing a baseline performance level.
Baselines are typically generated independently for each dataset using very simple models. Their role is to set the minimum level of acceptable performance and help with comparing relative improvements in performance of other models.
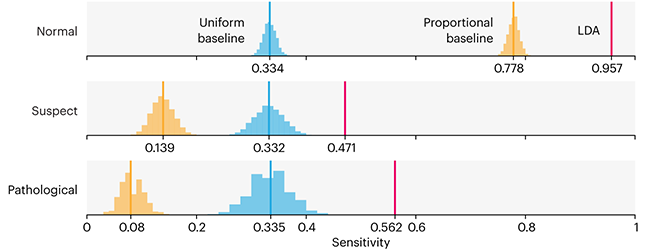
Unfortunately, baselines are often overlooked and, in the presence of a class imbalance5, must be established with care.
Megahed, F.M, Chen, Y-J., Jones-Farmer, A., Rigdon, S.E., Krzywinski, M. & Altman, N. (2024) Points of significance: Comparing classifier performance with baselines. Nat. Methods 20.
Happy 2024 π Day—
sunflowers ho!
Celebrate π Day (March 14th) and dig into the digit garden. Let's grow something.
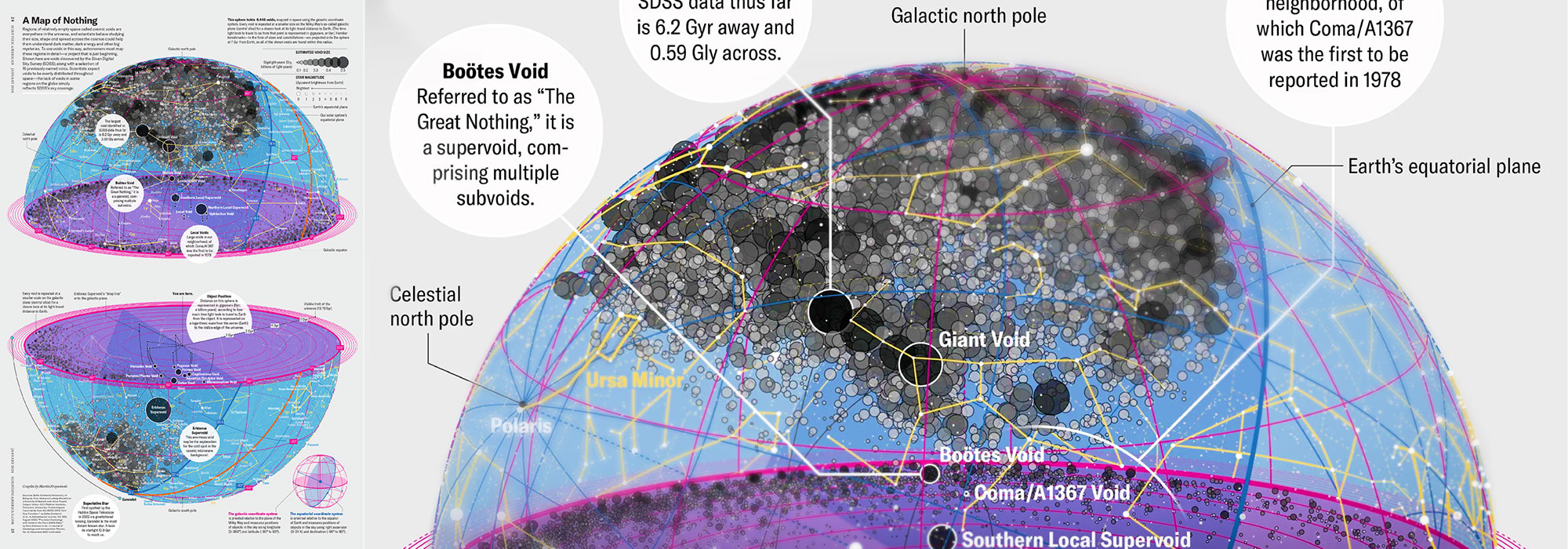
How Analyzing Cosmic Nothing Might Explain Everything
Huge empty areas of the universe called voids could help solve the greatest mysteries in the cosmos.
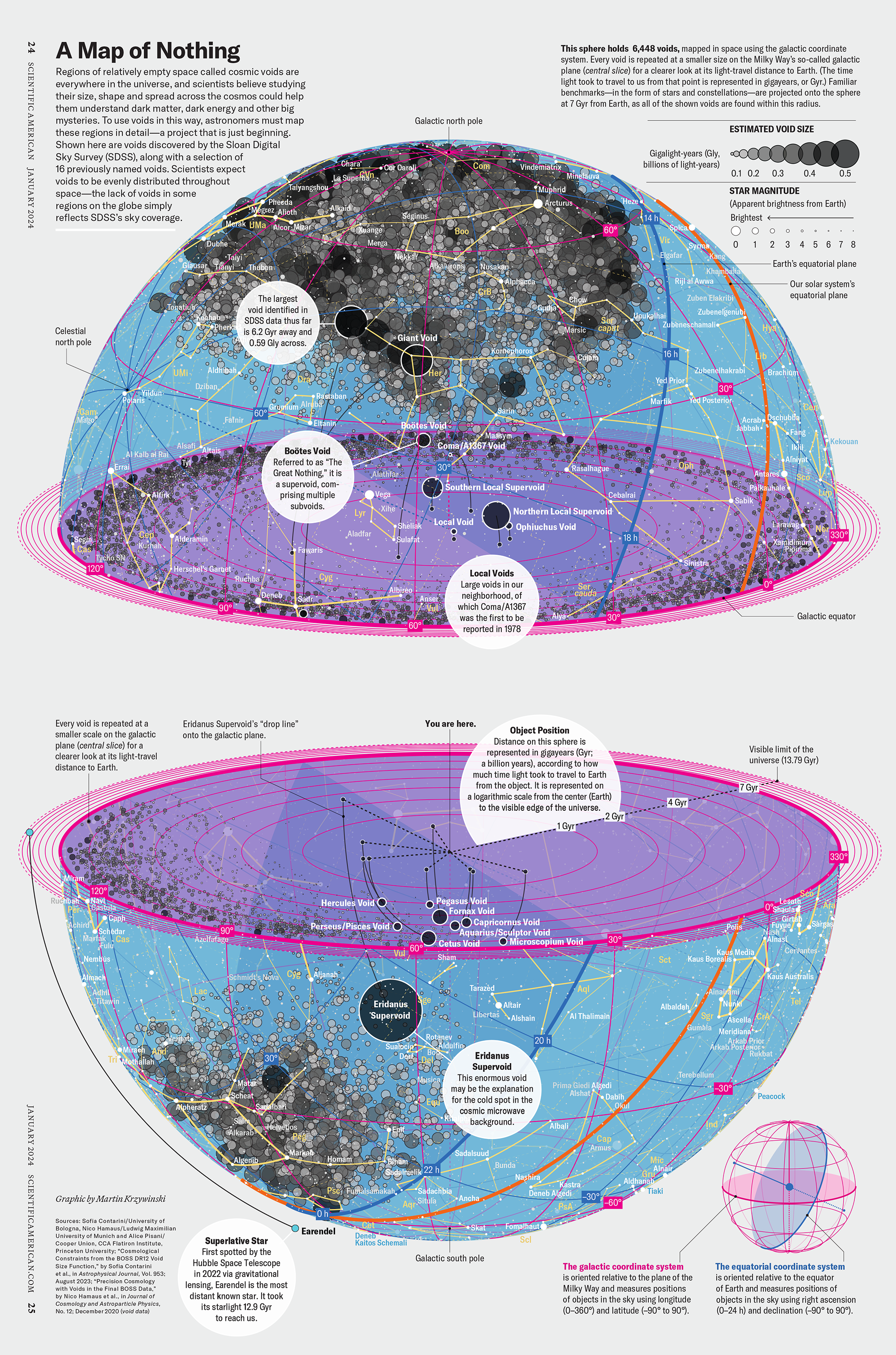
My graphic accompanying How Analyzing Cosmic Nothing Might Explain Everything in the January 2024 issue of Scientific American depicts the entire Universe in a two-page spread — full of nothing.
The graphic uses the latest data from SDSS 12 and is an update to my Superclusters and Voids poster.
Michael Lemonick (editor) explains on the graphic:
“Regions of relatively empty space called cosmic voids are everywhere in the universe, and scientists believe studying their size, shape and spread across the cosmos could help them understand dark matter, dark energy and other big mysteries.
To use voids in this way, astronomers must map these regions in detail—a project that is just beginning.
Shown here are voids discovered by the Sloan Digital Sky Survey (SDSS), along with a selection of 16 previously named voids. Scientists expect voids to be evenly distributed throughout space—the lack of voids in some regions on the globe simply reflects SDSS’s sky coverage.”
voids
Sofia Contarini, Alice Pisani, Nico Hamaus, Federico Marulli Lauro Moscardini & Marco Baldi (2023) Cosmological Constraints from the BOSS DR12 Void Size Function Astrophysical Journal 953:46.
Nico Hamaus, Alice Pisani, Jin-Ah Choi, Guilhem Lavaux, Benjamin D. Wandelt & Jochen Weller (2020) Journal of Cosmology and Astroparticle Physics 2020:023.
Sloan Digital Sky Survey Data Release 12
Alan MacRobert (Sky & Telescope), Paulina Rowicka/Martin Krzywinski (revisions & Microscopium)
Hoffleit & Warren Jr. (1991) The Bright Star Catalog, 5th Revised Edition (Preliminary Version).
H0 = 67.4 km/(Mpc·s), Ωm = 0.315, Ωv = 0.685. Planck collaboration Planck 2018 results. VI. Cosmological parameters (2018).
constellation figures
stars
cosmology
Error in predictor variables
It is the mark of an educated mind to rest satisfied with the degree of precision that the nature of the subject admits and not to seek exactness where only an approximation is possible. —Aristotle
In regression, the predictors are (typically) assumed to have known values that are measured without error.
Practically, however, predictors are often measured with error. This has a profound (but predictable) effect on the estimates of relationships among variables – the so-called “error in variables” problem.
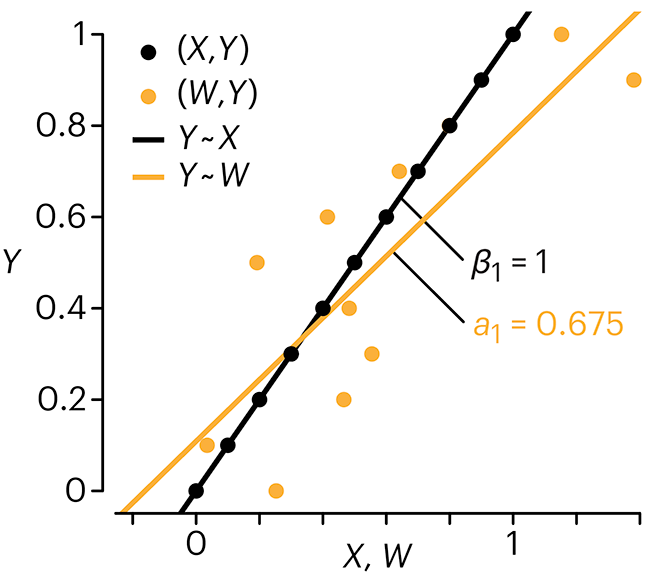
Error in measuring the predictors is often ignored. In this column, we discuss when ignoring this error is harmless and when it can lead to large bias that can leads us to miss important effects.
Altman, N. & Krzywinski, M. (2024) Points of significance: Error in predictor variables. Nat. Methods 20.
Background reading
Altman, N. & Krzywinski, M. (2015) Points of significance: Simple linear regression. Nat. Methods 12:999–1000.
Lever, J., Krzywinski, M. & Altman, N. (2016) Points of significance: Logistic regression. Nat. Methods 13:541–542 (2016).
Das, K., Krzywinski, M. & Altman, N. (2019) Points of significance: Quantile regression. Nat. Methods 16:451–452.
Convolutional neural networks
Nature uses only the longest threads to weave her patterns, so that each small piece of her fabric reveals the organization of the entire tapestry. – Richard Feynman
Following up on our Neural network primer column, this month we explore a different kind of network architecture: a convolutional network.
The convolutional network replaces the hidden layer of a fully connected network (FCN) with one or more filters (a kind of neuron that looks at the input within a narrow window).
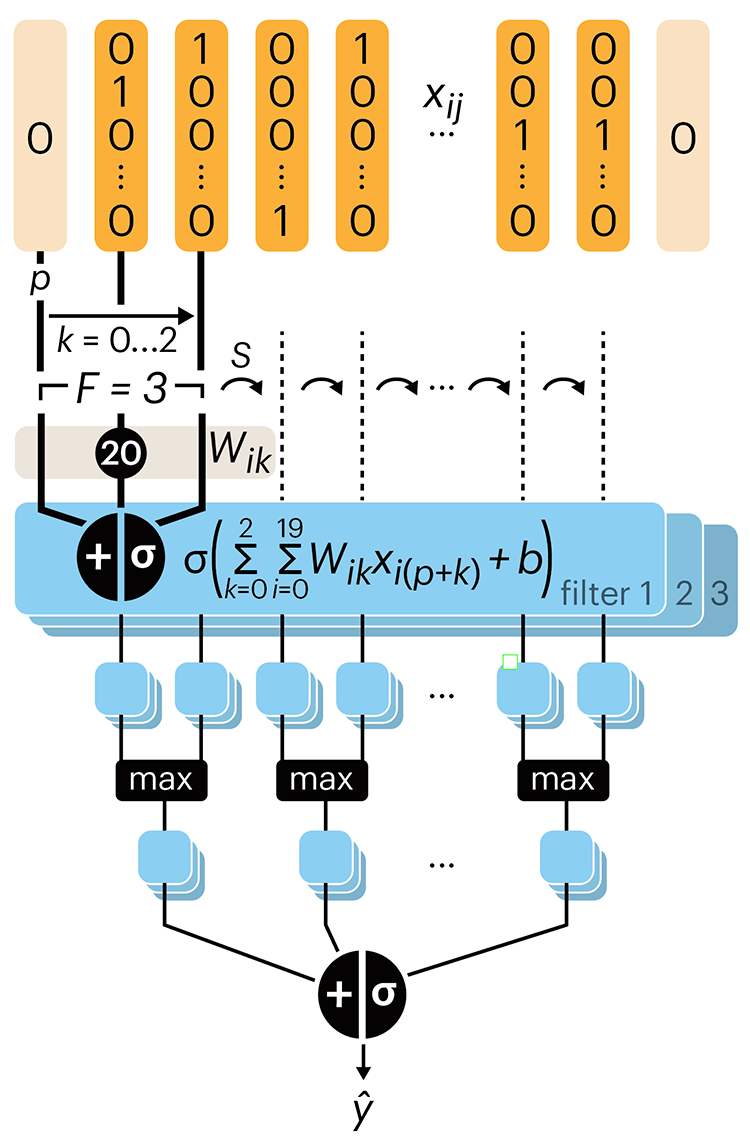
Even through convolutional networks have far fewer neurons that an FCN, they can perform substantially better for certain kinds of problems, such as sequence motif detection.
Derry, A., Krzywinski, M & Altman, N. (2023) Points of significance: Convolutional neural networks. Nature Methods 20:1269–1270.
Background reading
Derry, A., Krzywinski, M. & Altman, N. (2023) Points of significance: Neural network primer. Nature Methods 20:165–167.
Lever, J., Krzywinski, M. & Altman, N. (2016) Points of significance: Logistic regression. Nature Methods 13:541–542.